by Dennis Crouch
I have been following the ongoing case of Natera v. NeoGenomics that is currently on appeal before the Federal Circuit. Docket No. 24-1324. The technology in these cases has amazing potential and I have several friends who have used these techniques to detect early stage cancer.
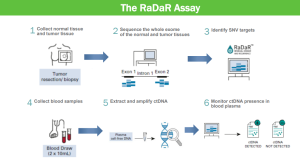
The particular litigation here centers on Natera's US Patent No. 11,519,035 issued on December 6, 2022 that covers a method of detecting cancer through analysis of cell-free DNA (cfDNA) obtained from blood samples. The basic idea is that small fragments of DNA that are released into the bloodstream by cells, including cancer cells. High throughput extraction and sequencing technologies can then be used to detect mutations and their potential associated risk factors, including the presence of cancer and the specific type of cancer. cfDNA cancer screening has lots of advantages, most notably, it is almost non-invasive (blood sample) and provides potential early across the entire body. It can be particularly effective for detecting reemergence of cancer for someone in remission because the particular mutation is already known. This is known as a "tumor-informed" test.
Natera's '035 patent is directed to methods for amplifying and sequencing cell-free DNA (cfDNA) to detect cancer. The claimed invention involves three key steps: (1) tagging cfDNA with universal adaptors, (2) amplifying 25-2,000 single nucleotide polymorphism (SNP) loci in a single reaction volume, and (3) performing massively parallel sequencing. Each of these steps were known in the prior art, but the combination of all three appears to be new. NeoGenomics is accused of infringing through its use of its RaDaR, an independently developed cancer-detection test that purportedly employs a similar process for analyzing cfDNA to provide highly sensitive detection of cancerous mutations. NeoGenomics disagrees, arguing the claims require the tagging and targeted amplification to occur in separate PCR reactions with different primer sets, and that its approach combines this process.
To continue reading, become a Patently-O member. Already a member? Simply log in to access the full post.